PHi-C2
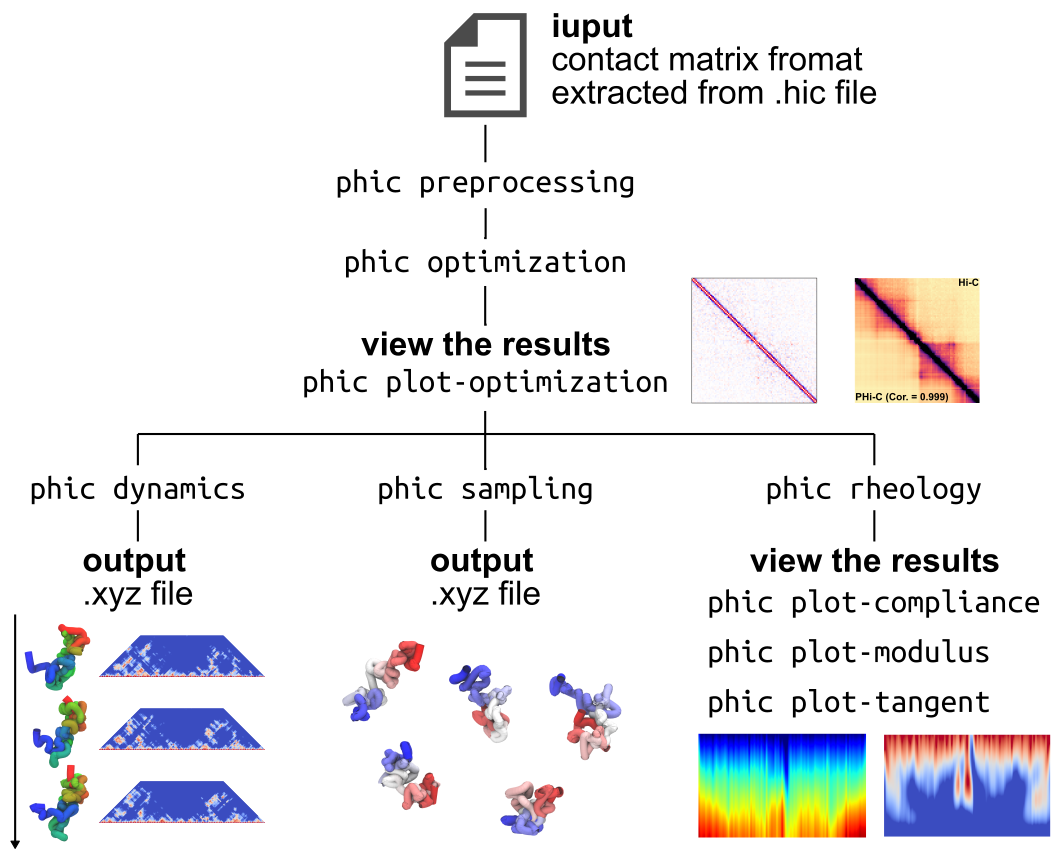
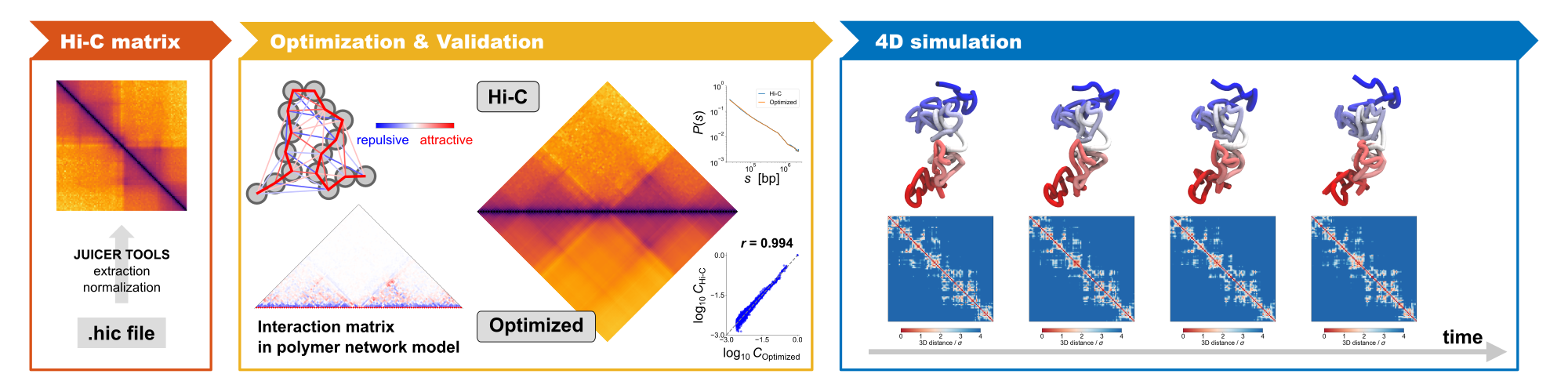
PHi-C2 is a Python package supported by mathematical and biophysical polymer modeling, that converts an input Hi-C matrix data into the polymer model's dynamics, structural conformations, and rheological features.
PHi-C2 as the phic package (maintaind on GitHub) is freely available and can be installed from the Python package index.
The updated optimization algorithm to regenerate a highly similar Hi-C matrix provides a fast and accurate optimal solution compared to the previous version by eliminating a computational bottleneck in the iterative optimization process.
In addition, without introducing a Python environment in the user's local platform, PHi-C2 runs on Google Colab.
Users can easily change parameters and check the results in the notebook.
Additionally, we shipped a command-line interface for convenient application.