How is the genome organized in living cells?
The genome is not only one-dimensional information of DNA sequences but also is packed and folded three-dimensionally inside the cell nucleus, which is only a few microns in size. In the micro-scale world, thermal fluctuations are inevitable and cause random and stochastic motion of all molecules in living cells. Our research interest is to know how the genome is organized and functional in living cells as the fluctuating world. What are the physical principles of the 3D genome organization? We combine theoretical and experimental approaches from biophysics with analytical methods from genomics to understand the nature of the dynamic 3D genome organization.
Dynamic movements of single-nucleosomes in living cells
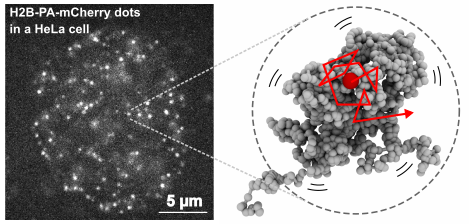
Chromatin is highly dynamic in the cell nucleus. My collaborator, Prof. Maeshima at NIG Japan, has revealed subdiffusively dynamic nucleosome movements in living mammalian cells using single-nucleosome imaging and found liquid-like irregular folding of chromatin. How is the dynamic movement regulated?
Snapshots of fragments of the 3D genome organization
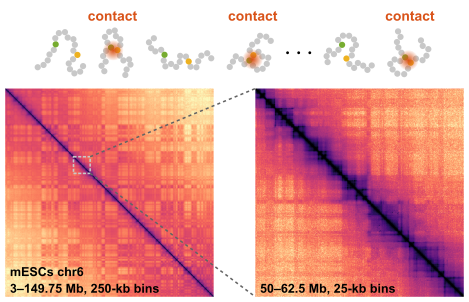
Hi-C technologies have uncovered the nature of the 3D chromatin folding. The Hi-C matrix consists of a massive amount of data about the contact DNA pairs' fragments in a plethora of fixed cells. The unique patterns imply characteristic 3D folding of chromatin fibers in the cell nuclei. Can we decode the static Hi-C data into the dynamic state of the genome?
Polymer modeling can bridge the dynamics and structure
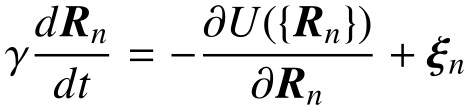
Chromatin is a polymeric fiber subject to thermal fluctuations. In polymer modeling of the chromatin, describing all interaction potential energies allows for theoretical analyses and simulations of the chromatin dynamics and structure. What is a set of minimal physical mechanisms underlying the 3D genome organization? What are physical interactions to reproduce Hi-C matrix data? Can we predict the chromatin dynamics and structure in living cells?